
GANDALF
Genomic AND Adaptation for fungal LiFe history traits involved in host plant interaction
Parasites are able to evolve rapidly and overcome host defense mechanisms, but the molecular basis of this adaptation remains poorly understood. Identifying polymorphisms under selection in pathogenic fungal populations will help understanding the evolutionary processes underlying the adaptation to host-plant. Fungal genes may qualitatively determine whether a plant genotype can be infected or not (i.e. host specialization and/or virulence/avirulence) or may quantitatively determine the ability of the fungus to overcome the basal defense of the host plant species (i.e. aggressiveness). Recently, it has been proposed that the same molecular mechanisms could in fact underlie these seemingly different fungal traits. Taking advantage of the advance of Next Generation Sequencing technologies, GANDALF proposes to use a population genomics approach (i.e. without any a priori on the genes involved in host adaptation) for addressing the processes of adaptation in the ecological/agronomic gradient from host-plant specialization to quantitative adaptation to host-plant resistance. GANDALF is bringing together scientists from different laboratories and institutions having internationally recognized expertise in plant pathology, genomics, bioinformatics, evolutionary genetics and modelling (all ranked A by AERES, 4 being A+).
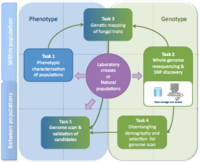
Nine different pathosystems have been chosen, covering a large range of life history traits (e.g. biotophic vs necrotrophic, fungi vs oomycetes, etc.) and with genome sequences already available. Depending on the system, offspring of controlled crosses or natural populations will be analyzed. A first step aims at characterising strains for traits associated with host adaptation (phenotyping, Task 1). Re-sequencing of phenotyped strains will allow assembling, mapping and SNP discovery (Task 2). Genome-wide polymorphism data will be analysed following either a genome-wide association (Task 3) or a genome scan approach (Task 5). Since pathogens are prone to demographic and spatial variations, a method able to infer demographic parameters on populations will be developed in order to build a powerful tool of selection detection under complex population models (Task 4). Using this new tool, genome scan will be performed in order to detect loci under selection (Task 5). The candidate loci identified in tasks 3 and 5 will be validated by further genotyping on natural populations or collections available in laboratories. The genetic bases of host adaptation will be compared among fungal species and along the host specialization / cultivar specificity / aggressiveness continuum.
This project not only will bring new insights into the understanding of genomic basis of fungal plant pathogens, but also will contribute to maintain the scientific excellence of the GANDALF partners by accelerating the integration of NGS technologies into their research programs.
Duration: 01/01/2013 to 31/12/2016
Coordinator: François Delmotte
Partners
- ESE, Paris-Orsay ( http://www.ese.u-psud.fr/ ), Tatiana Giraud
- BGPI, Montpellier ( http://umr-bgpi.cirad.fr/ ), Jean Carlier
- BIOGER, Paris-Grignon ( http://www.versailles-grignon.inra.fr/bioger ), Thierry Marcel
- SAVE, Bordeaux ( http://www6.bordeaux-aquitaine.inra.fr/sante-agroecologie-vignoble/ ), François Delmotte
- IAM, Nancy ( http://www.nancy.inra.fr/Le-centre-Les-recherches/Les-unites/IAM ), Fabien Halkett
- BIOGECO, Bordeaux ( https://www4.bordeaux-aquitaine.inra.fr/biogeco/ ), Cyril Dutech
- IRHS, Angers ( http://www6.angers-nantes.inra.fr/irhs ), Bruno Lecam
- URGI ( http://urgi.versailles.inra.fr/ ), Joelle Amselem
- LIPM, Toulouse ( http://www6.toulouse.inra.fr/lipm ), JĂ©rĂ´me Gouzy
Project funded by ANR BIOADAPT
Creation date: 20 Jun 2013
 eZ Publish
eZ PublishPublication supervisor: A-F. Adam-Blondon
Read Credits & General Terms of Use
Read How to cite









