
VRE Epigenetics
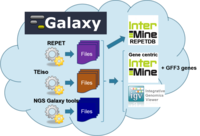
URGI in-house project to set up a Virtual Research Environment to study Transposable Elements and their impact on the epigenomes.
The goal is to build a research environment using:
- Analysis tools
- Visualization tools
- Data sharing tools
- Virtual Machines
Analysis tools:
- Repeat analysis: REPET , TEiso
- Read mapping: BWA
- NGS Galaxy tools
- RNASeq and ChiPseq analysis
Visualization tools:
- Genomes (genes, TE, RNAseq, etc.): IGV
- TE consensus: IGV, Jalview
Data sharing tools:
- Files sharing: Fuse, FTP/SSH, iRODS
- Files indexation: ElasticSearch
- Databases: InterMine ( RepetDB ), TripleStore
Virtual Machines:
- These tools will be installed on the Galaxy environment in Virtual Machines.
- VMs allow easy copy and distribution.
Update: 17 Feb 2017
Creation date: 13 Feb 2017
Creation date: 13 Feb 2017
QUALITÉ
ANNOTATION
DATA SERVICES
PLATFORM
QUALITY
PROJECTS
DATA
TOOLS
SPECIES
ABOUT US
CONTACT US
REGISTER
EDIT
Copyright © INRAE 2020 - Designed by SWAD, developed by URGI with  eZ Publish
eZ Publish
Publication supervisor: A-F. Adam-Blondon
Read Credits & General Terms of Use
Read How to cite
 eZ Publish
eZ PublishPublication supervisor: A-F. Adam-Blondon
Read Credits & General Terms of Use
Read How to cite








