
Caulifinder README
CAULIFINDER

QUICK START
The CAULIFINDER image and instructions are available on docker hub: https://hub.docker.com/r/urgi/docker_caulifinder
OVERVIEW
CAULIFINDER was developed to facilitate the discovery and annotation of endogenous viral elements (EVEs) of caulimovirid origin in plant genomes. CAULIFINDER allows quantifying the contribution of caulimovirid EVEs to any plant genome, addressing their impact on plant biology and collecting sequences that can add to paleovirology studies.
CAULIFINDER consists of two complementary search strategies available as two distinct pipelines. The first one, branch A, is used to reconstruct consensus sequences that are representative of repetitive caulimovirid EVE copies and to annotate plant genome assemblies accordingly. The second one, branch B, scans plant genomes and transcriptomes for caulimovirid reverse transcriptase (RT) domains and it include representative sequences in a phylogenetic tree to address their diversity.
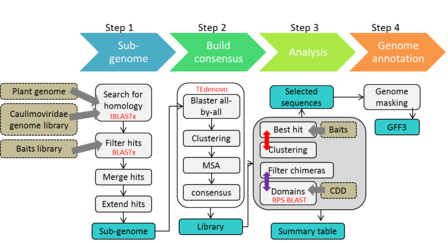
The BRANCH A pipeline consists of four steps and produces the following main output files:
- A library of consensus sequences (fasta format)
- A table summarizing the main features of each consensus (gff format)
- An annotation file listing the positions of caulimovirid EVEs in the genome of interest (table)
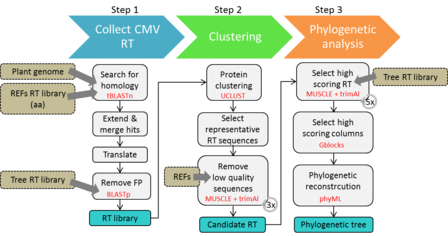
The BRANCH B pipeline comprises three steps and generates the following key output files:
- A library of representative RT sequences (fasta format)
- A multiple sequence alignment for RT (fasta format)
- A phylogenetic tree (Newick format)
The branch A&B pipelines are described in details in the dedicated publication (https://doi.org/10.1186/s13100-022-00288-w)
AVAILABILITY
CAULIFINDER pipelines are versioned as a gitlab project saved in forgemia . The fasta libraries and the documentation to launch Branch A and Branch B are also available in this gitlab project. The fasta libraries are also published in a dataverse where they will be updated and versioned.
For the ease of installation, the CAULIFINDER container is distributed as a docker image which is available on docker hub here: https://hub.docker.com/r/urgi/docker_caulifinder .
The container code is deposited in the public gitlab repository Caulifinder _ docker
! Due to the usage of MariaDB, you will need administrator rights to run the docker image!
README
The documentation to use the docker image and the full CAULIFINDER usage description are also available in the public gitlab repository Caulifinder _ docker
SUPPORT
For any question, please contact florian.maumus@inrae.fr
CITATION
If you use CAULIFINDER, please cite
Vassilieff, H., Haddad, S., Jamilloux, V. et al. CAULIFINDER: a pipeline for the automated detection and annotation of caulimovirid endogenous viral elements in plant genomes. Mobile DNA 13, 31 (2022). https://doi.org/10.1186/s13100-022-00288-w
Creation date: 19 Feb 2024
 eZ Publish
eZ PublishPublication supervisor: A-F. Adam-Blondon
Read Credits & General Terms of Use
Read How to cite








