VRE Epigenetics
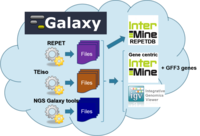
URGI in-house project to set up a Virtual Research Environment to study Transposable Elements and their impact on the epigenomes.
The goal is to build a research environment using:
- Analysis tools
- Visualization tools
- Data sharing tools
- Virtual Machines
Analysis tools:
- Repeat analysis: REPET , TEiso
- Read mapping: BWA
- NGS Galaxy tools
- RNASeq and ChiPseq analysis
Visualization tools:
- Genomes (genes, TE, RNAseq, etc.): IGV
- TE consensus: IGV, Jalview
Data sharing tools:
- Files sharing: Fuse, FTP/SSH, iRODS
- Files indexation: ElasticSearch
- Databases: InterMine ( RepetDB ), TripleStore
Virtual Machines:
- These tools will be installed on the Galaxy environment in Virtual Machines.
- VMs allow easy copy and distribution.