GnpInteGr
GnpIS
GnpInteGr is an ANR 2007 bioinformatics project, coordinated by INRA URGI, dedicated to the development on an interoperability between tools and data, existing locally in URGI databases or in external resources.
ANR-07-GPLA-005-001
ANR GnpInteGR project is a innovative bioinformatics development project that helps both fondamental and industrial genomic research by providing tools to scientists to help them in data mining, for example in finding genes of agronomical interest, linked to QTLs or to find allèles of interest for breeders. It helps scientists also in the setting of new biological experiments to confirm hypothesis and in silico analysis for example in gene function prediction. It promotes the starting of new project deposits at national and international level and contribute to the valorization of both tools and data produced.
The project was set up in 2008 to improve the existing information system GnpIS , which was itself developped in collaboration with INRA public and private (Biogemma, Bayer) partners through mainly fundings allocated year after year, since 2000 in the frame of Genoplante calls (Genoplante Bioinformatics transversal, BIEP, M2 projects) . ANR GnpInteGr was written for 36 monthes and one year was funded to hire one CDD developper, Aminah-Olivia Keliet and to add new hardware equipments.
Duration: 01/01/2008 to 30/09/2009
Coordinator: Delphine Steinbach
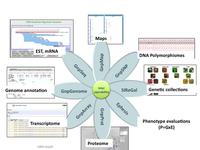
GnpIS architecture, a web based system composed of 7 relational databases accessible via a unique portal (http://urgi.versailles.inra.fr/gnpis)
GnpIS URGI information system relies on cutting edge databases and data warehouse technologies. It is a web based system composed of 7 relational databases. The key feature is that they are connected through the sharing of common tables and cross-reference links and it relies on a powerful, modular and extensible software architecture. GnpIS modules are built using the Service-Oriented Architecture (SOA), which is an architectural approach for constructing complex software-intensive systems from a set of universally interconnected and interdependent building blocks, called services. The platform chosen for the implementation of the SOA is the open source J2EE platform (Sun Microsystems). GnpIS/GnpGenome annotation sequence module use GMOD (chado/gbrowse) open source tools.
The technologies that were chosen in this project to query all the databases modules at the same time, were i) the use of Lucene, a high-performance, full-featured text search engine and especially HibernateSearch, that brings the power of full text search engines to the persistence domain model and the use of 2) Biomart, an open source query-oriented data management system.
Major results and deliverables since 2009:
- A new portal, https://urgi.versailles.inra.fr/gnpis , which relies on existing local databases.
- 2 new tools to query through all these databases transparently.
- The first tool, called “quick search” is based on Lucene technologies. It allows to search data in GnpIS indexes, as a ‘google search’ and to get results as clickable item lists to go deeper in information.
- The second tool called, ‘advanced search tool ’, is based on Biomart and on new datamarts that we created from GnpIS. It allows to query precomputed data by selecting, combining and filling predefined fields.
- The system was instantiated with 3 sets of real data for Wheat, Grapevine and Poplar data .
Communication:
Publication in prep. in 2010:
- GnpIS: an original information system to bridge genetic and genomic plant and fungi data: Delphine Steinbach, Erik Kimmel, Aminah-Olivia Keliet, Daphné Verdelet, Michael Alaux, Nathalie Choisne, Joelle Amselem, Nacer Mohellibi, Isabelle Luyten, Sophie Durand, Cyril Pommier, Sébastien Reboux, Hadi Quesneville
Posters :
- GnpIS update: poster and oral session for ANR GnpInteGR project by Delphine Steinbach, at Genoplante annual seminar in 2010.
- GnpIS update: two new query tools to bridge plant genetics and genomics data - D. Steinbach, E. Kimmel, A.-O. Keliet, M. Alaux, J. Amselem, S. Durant, C. Pommier, I. Luyten, N. Mohellibi, D. Verdelet, S. Reboux, H. Quesneville, Plant Genomics European Meetings, October 7-10, 2009, Lisbon, Portugal.
- GnpIS: Focus on wheat genomic data - M. Alaux, D. Verdelet, A.-O. Keliet, E. Kimmel, B. Brault, N. Mohellibi, B. Hilselberger, S. Durant, I. Luyten, S. Reboux, D. Steinbach, Plant Genomics European Meetings, October 7-10, 2009, Lisbon, Portugal.
- GnpIS, the plant information system of INRA URGI bioinformatics platform - D.Steinbach, M.Alaux, E.Kimmel, S.Derozier, S.Durand, C.Pommier, I.Luyten, N.Mohellibi, D.Verdelet, S.Reboux and H.Quesneville, PAG XVII Jan 2009, San Diego, USA
Talks or demo:
- Project report on GnpIS and its new query tools, by Delphine Steinbach, at Genoplante annual seminar in 2010.
- The URGI bioinformatic platform: an original information system to bridge genetic and genomic plant and fungal data " at ISYIP 2009, invited speaker (Hadi Quesneville) at The 1st International Workshop on "Information SYstems for Insect Pests" (ISYIP), INRIA, Rennes, France.
- Wheat data on GnpIS, the INRA URGI plant information system - M. Alaux , D. Steinbach, E. Kimmel, S. Durand, C. Pommier, N. Mohellibi, D. Verdelet, I. Luyten, S. Reboux, H. Quesneville . 19th International Triticeae Mapping Initiative - 3rd COST Tritigen, August 31th - September 4th 2009, Clermont-Ferrand, France.
- Project report on GnpIS and its new query tools, by Delphine Steinbach, at INRA URGI platform annual CSU meeting.
- PAG 2009, URGI demo and posters : The online demo of "GnpIS, the plant information system of INRA URGI bioinformatics platform" presented at PAG 2009 San Diego by Michael Alaux
- Genomic and genetic data integration : The high throughput challenge - D.Steinbach, M.Alaux, J.Amselem, S.Durand, E.Kimmel, I.Luyten, N.Mohellibi, C.Pommier, S.Reboux, H.Quesneville (speaker), SFG and SFGH meeting: "Genetics and Epigenetics. New Approaches in Genomics", 22nd and 23rd Jan 2009, Pasteur Institute, France
Training Sessions:
- Training on GnpIS and especially on its 2 new query tools:
- At INRA Clermont (GDEC unit) for wheat genomics
- At INRA Montpellier (DIA PC unit) for grape genomics
- At INRA Colmar for grape genomics